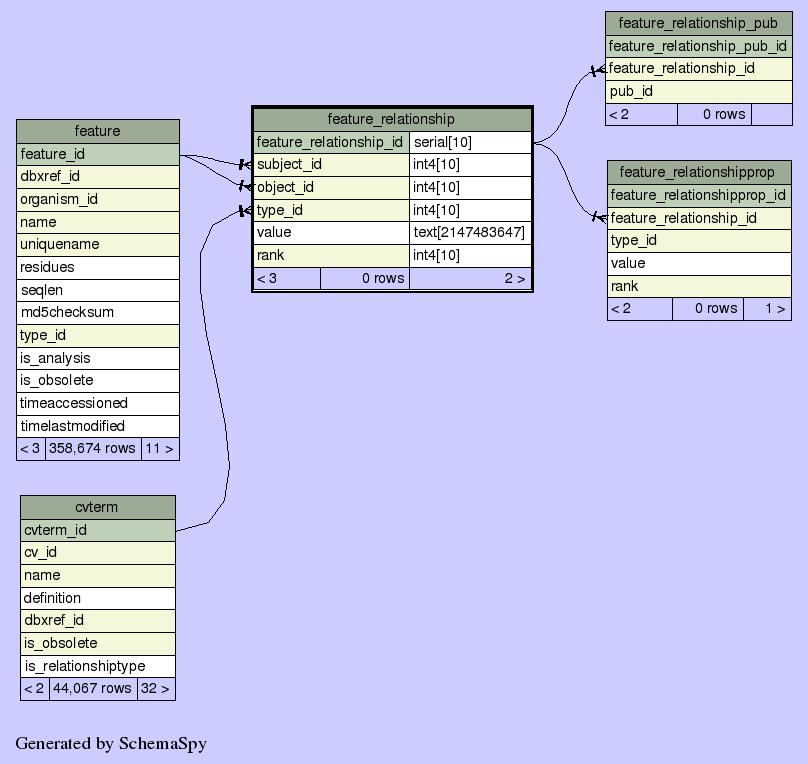
Table cxgn.public.feature_relationshipfeatures can be arranged in
graphs, eg exon part_of transcript part_of gene; translation madeby transcript if type is thought of as a verb, each arc makes a statement [SUBJECT VERB OBJECT] object can also be thought of as parent (containing feature), and subject as child (contained feature or subfeature) -- we include the relationship rank/order, because even though most of the time we can order things implicitly by sequence coordinates, we cant always do this - eg transpliced genes. its also useful for quickly getting implicit introns |
Generated by SchemaSpy |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Indexes:
| Column(s) | Type | Sort | Constraint Name |
|---|---|---|---|
| feature_relationship_id | Primary key | Asc | feature_relationship_pkey |
| subject_id + object_id + type_id + rank | Must be unique | Asc/Asc/Asc/Asc | feature_relationship_c1 |
| subject_id | Performance | Asc | feature_relationship_idx1 |
| object_id | Performance | Asc | feature_relationship_idx2 |
| type_id | Performance | Asc | feature_relationship_idx3 |
|