Stack/Anderson Lab

Front Row (left to right): Dr. Lorrie Anderson
(assistant professor), Ann Lai (research associate), Jia Cheng
(graduate student), Stephanie Lum (undergraduate). Back
Row (left to right): Joe Qiao (graduate student), Kevin
Su (undergraduate), Dr. Stephen Stack (professor), Suzanne Royer
(research associate), Erin Benson (undergraduate), Song-Bin Chang
(post doc). Absent from photo: Brittany Howard
(undergraduate).
We work primarily with two proteinaceous structures: the synaptonemal complex (SC) and recombination nodules (RNs). The SC looks like a railroad track that is formed between synapsed homologous chromosomes, and RNs are 100 nm particles that occur on SCs at sites where crossing over occurs and where chiasmata will form later. We pioneered a technique for spreading complete sets of plant SCs for analysis by electron microscopy. We use this technique to determine the pattern and frequency of crossing over in wild type plants and in plants with chromosome aberrations such as translocations and inversions.

Fluorescence in situ hybridization (FISH) on spreads of tomato
synaptonemal complexes.
We are particularly interested in the relation between genes and chromosome structure and the physical relation of recombination proteins to SCs and RNs. Ultimately we would like to determine the roles of SCs and RNs in crossing over and interference.
This is an exciting time in cytogenetics with so much new information, so many interesting questions, and so many new techniques and instruments to help find the answers. All that must be added for continued progress is imagination and hard work.
Currently we are involved in the following projects:
- Fluorescence in situ hybridization (FISH) on spreads of tomato synaptonemal complexes (SCs = pachytene chromosomes) to determine the physical location of tomato DNA inserts in bacterial artificial chromosomes (BACs) (See chromosome image above.). FISH is integral to the tomato genome sequencing project 1) to keep the sequencing effort confined primarily to euchromatin, 2) to locate problem BACs, and 3) to define the size of gaps in chromosome walks.
- Light and electron microscopic immunolocalization of proteins thought to be involved in recombination. This work concentrates on the timing of the appearance and disappearance of these proteins in relation to the SC and recombination nodules (RNs). See figure below.
- Light and electron microscopic characterization of chiasmata and RNs in maize mutants that show changes in the rate and/or location of crossing over.
- Spreading SCs from both plants and mammals to characterize the effects of chromosome aberrations such as translocations and inversions on the number and distribution of crossover events as indicated by RNs.
Contact Information
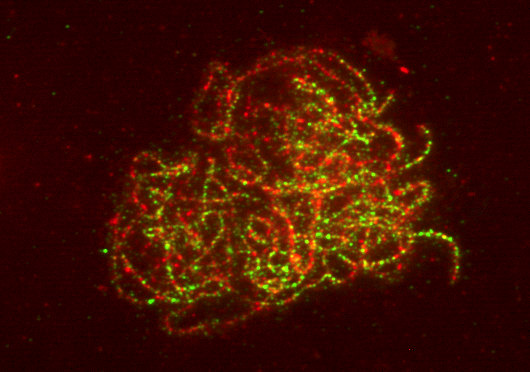
Tomato immunolabeling: The photo is of a tomato spread (early
zygotene, as the chromosomes are beginning to synapse)
immunolabeled with Mre11 (green spots) and Smc1 (red). Mre11 is a
DNA double-strand break repair protein and Smc1 is labeling the
chromosome core (therefore enabling us to see the synaptonemal
complex under fluorescent microscopy).
Stephen Stack
Department of Biology
Colorado State University
Fort Collins, Colorado 80523-1878
USA
Telephone: 970-491-6802
FAX: 970-491-0649
E-mail sstack@lamar.colostate.edu
Lorinda Anderson
Department of Biology
Colorado State University
Fort Collins, Colorado 80523
USA
Telephone: 970-491-4856
FAX: 970-491-0649
E-mail: lorrie@lamar.colostate.edu
Selected Publications
Sherman, J.D. and S. M. Stack. 1995. Two-dimensional spreads of synaptonemal complexes from solanaceous plants. VI. High resolution recombination nodule map for tomato (Lycopersicon esculentum). Genetics 141:683-708
Peterson, D.G., H.J. Price, J.S. Johnston, and S.M. Stack. 1996. DNA content of heterochromatin and euchromatin in tomato (Lycopersicon esculentum) pachytene chromosomes. Genome 39:77-82
Peterson, D.G., K.S. Boehm, and S.M. Stack. 1997. Isolation of milligram quantities of nuclear DNA from tomato (Lycopersicon esculentum), a plant containing high levels of polyphenolic compounds. Plant Molec. Biol. Reporter 15:148-153
Peterson, D.G., W.R. Pearson, S.M. Stack. 1998. Characterization of the tomato (Lycopsersicon esculentum) genome using in vitro and in situ DNA reassociation. Genome 41:346-356
Peterson, D.G., N.L.V. Lapitan, and S.M. Stack. 1999. Localization of single- and low-copy sequences on tomato synaptonemal complex spreads using fluorescence hybridization (FISH). Genetics 152:427-439
Stack, S.M. and L.K. Anderson. 2001. A model for chromosome structure during the mitotic and meiotic cell cycles. Chromosome Research 9:175-198
Anderson, L.K. and S.M. Stack 2001. Distribution of early recombination nodules on zygotene bivalents from plants. Genetics 159:1259-1269
Anderson, L.K., K.D. Hooker, and S.M. Stack 2001. The distribution of early recombination nodules on zygotene bivalents from plants. Genetics 159:1259-1269
Stack, S.M. and L.K. Anderson 2002. Crossing over as assessed by late recombination nodules is related to the pattern of synapsis and the distribution of early recombination nodules in maize. Chromosome Research 10:329-345
Anderson, L.K. and S.M. Stack 2002. Meiotic recombination in plants. Current Genomics 3:507-526
Tenaillon, M.I., M.C. Sawkins, L.K. Anderson, S.M. Stack, J. Doebley, and B.S. Gaut 2002. Patterns of diversity and recombination along chromosome 1 of maize (Zea mays ssp. Mays L.) Genetics 162:1401-1413
Anderson, L.K., G.C. Doyle, B. Brigham, J. Carter, K.D. Hooker, A. Lai, M. Rice, and S.M. Stack. 2003. High resolution Crossover maps for each bivalent of Zea mays using recombination nodules. Genetics 165:849-865.
Anderson, L.K., N. Salameh, H.W. Bass, L.C. Harper, W.Z. Cande, G. Weber, and S.M. Stack. 2004. Integrating genetic linkage maps with pachytene chromosome structure in maize. Genetics 166:1923-1933.



