This site uses cookies to provide logins and other features. Please accept the use of cookies by clicking Accept.
Tomato locus notabilis
| Locus details | Download GMOD XML | Note to Editors | Annotation guidelines |
[loading edit links...]
|
[loading...]
|
|
 Links to external databases Links to external databases
 Links to external databases Links to external databases
|
 Registry name: ABA deficient Registry name: ABA deficient
 Registry name: ABA deficient Registry name: ABA deficient
| [Associate registry name] |
This locus is associated with registry name: ABA deficient
 Notes and figures (2) Notes and figures (2)
 Notes and figures (2) Notes and figures (2)
| [Add notes, figures or images] |
 Accessions and images (7) Accessions and images (7)
 Accessions and images (7) Accessions and images (7)
| [Associate accession] |
 Alleles (1) Alleles (1)
 Alleles (1) Alleles (1)
| [Add new Allele] |
 Associated loci (1) Associated loci (1)
 Associated loci (1) Associated loci (1)
| [Associate new locus] |
[loading...]
|
 Associated loci - graphical view Associated loci - graphical view
 Associated loci - graphical view Associated loci - graphical view
| View notabilis relationships in the stand-alone network browser |
[loading...] | [Legend] [Levels] |
 SolCyc links SolCyc links
 SolCyc links SolCyc links
|
[loading...]
 Sequence annotations Sequence annotations
 Sequence annotations Sequence annotations
|
 Genome features Genome features
 Genome features Genome features
|
 Genomic sequence Genomic sequence
 Genomic sequence Genomic sequence
| unprocessed genomic sequence region underlying this gene |
>Solyc07g056570.1 SL2.50ch07:64363163..64361346 (sequence from reverse strand)
ATGGCAACTACTACTTCACATGCCACAAATACATGGATTAAGACTAAGTTGTCAATGCCATCATCAAAGGAGTTTGGTTTTGCATCAAACTCTATTTCTCTACTCAAAAATCAACATAATAGGCAAAGTCTCAACATTAATTCCTCTCTTCAAGCTCCACCTATACTTCATTTTCCTAAACAATCTTCAAATTATCAAACACCAAAGAATAATACAATTTCACACCCAAAACAAGAAAACAACAACTCCTCTTCTTCTTCAACTTCCAAGTGGAATTTAGTGCAGAAAGCAGCAGCAATGGCTTTAGATGCTGTAGAAAGTGCTTTAACTAAACATGAACTTGAACACCCTTTGCCGAAAACAGCCGACCCACGAGTCCAGATTTCTGGGAATTTTGCTCCGGTACCGGAAAATCCAGTCTGTCAATCTCTTCCGGTCACCGGAAAAATACCCAAATGTGTTCAAGGCGTTTACGTTCGAAACGGAGCTAACCCTCTTTTTGAACCAACCGCCGGACACCATTTCTTCGACGGCGACGGTATGGTTCACGCCGTTCAATTCAAAAATGGGTCGGCTAGTTACGCTTGCCGTTTCACTGAAACAGAGAGGCTTGTTCAAGAAAAAGCTTTGGGTCGCCCTGTTTTCCCTAAAGCCATTGGTGAATTACATGGTCACTCTGGAATTGCAAGGCTTATGCTGTTTTACGCTCGTGGGCTCTTCGGACTTGTTGATCACAGTAAAGGAACTGGTGTTGCAAACGCCGGTTTAGTCTATTTCAATAACCGATTACTTGCTATGTCTGAAGATGATTTGCCTTACCATGTAAAGGTAACACCCACCGGCGATCTTAAAACAGAGGGTCGATTCGATTTCGACGGCCAGCTAAAATCCACCATGATAGCTCACCCAAAGCTCGACCCAGTTTCCGGTGAGCTATTTGCTCTTAGCTACGATGTGATTCAGAAGCCATACCTCAAGTACTTCAGATTTTCAAAAAATGGGGAAAAATCAAATGATGTTGAAATTCCAGTTGAAGACCCAACAATGATGCATGATTTCGCAATTACTGAGAACTTCGTCGTCATTCCTGATCAACAAGTCGTTTTCAAGATGTCTGAAATGATCCGTGGAGGTTCACCGGTGGTTTACGACAAGAACAAAGTTTCCCGATTTGGTATTCTGGATAAGTACGCGAAAGATGGGTCTGATTTGAAATGGGTTGAAGTACCTGATTGTTTCTGTTTCCACCTCTGGAATGCTTGGGAAGAAGCAGAAACAGATGAAATCGTTGTAATTGGTTCATGTATGACACCACCAGACTCCATTTTCAATGAATGTGATGAAGGGCTAAAGAGTGTTTTATCCGAAATCCGTCTCAATTTGAAAACAGGGAAATCAACAAGAAAATCCATAATCGAAAACCCGGATGAACAAGTGAATTTAGAAGCTGGAATGGTGAACCGAAACAAACTCGGAAGGAAAACAGAGTATGCTTATTTGGCTATCGCTGAACCATGGCCAAAAGTTTCTGGTTTTGCAAAAGTAAACCTGTTCACCGGTGAAGTTGAGAAATTCATTTATGGTGACAACAAATATGGTGGGGAACCTCTTTTTTTACCAAGAGACCCCAACAGCAAGGAAGAAGACGATGGTTATATTTTAGCTTTCGTTCACGATGAGAAAGAATGGAAATCAGAACTGCAAATTGTTAACGCAATGAGTTTGAAGTTGGAGGCAACTGTGAAGCTTCCATCAAGAGTTCCTTATGGATTTCATGGAACATTCATAAACGCCAATGATTTGGCAAATCAGGCATGA
ATGGCAACTACTACTTCACATGCCACAAATACATGGATTAAGACTAAGTTGTCAATGCCATCATCAAAGGAGTTTGGTTTTGCATCAAACTCTATTTCTCTACTCAAAAATCAACATAATAGGCAAAGTCTCAACATTAATTCCTCTCTTCAAGCTCCACCTATACTTCATTTTCCTAAACAATCTTCAAATTATCAAACACCAAAGAATAATACAATTTCACACCCAAAACAAGAAAACAACAACTCCTCTTCTTCTTCAACTTCCAAGTGGAATTTAGTGCAGAAAGCAGCAGCAATGGCTTTAGATGCTGTAGAAAGTGCTTTAACTAAACATGAACTTGAACACCCTTTGCCGAAAACAGCCGACCCACGAGTCCAGATTTCTGGGAATTTTGCTCCGGTACCGGAAAATCCAGTCTGTCAATCTCTTCCGGTCACCGGAAAAATACCCAAATGTGTTCAAGGCGTTTACGTTCGAAACGGAGCTAACCCTCTTTTTGAACCAACCGCCGGACACCATTTCTTCGACGGCGACGGTATGGTTCACGCCGTTCAATTCAAAAATGGGTCGGCTAGTTACGCTTGCCGTTTCACTGAAACAGAGAGGCTTGTTCAAGAAAAAGCTTTGGGTCGCCCTGTTTTCCCTAAAGCCATTGGTGAATTACATGGTCACTCTGGAATTGCAAGGCTTATGCTGTTTTACGCTCGTGGGCTCTTCGGACTTGTTGATCACAGTAAAGGAACTGGTGTTGCAAACGCCGGTTTAGTCTATTTCAATAACCGATTACTTGCTATGTCTGAAGATGATTTGCCTTACCATGTAAAGGTAACACCCACCGGCGATCTTAAAACAGAGGGTCGATTCGATTTCGACGGCCAGCTAAAATCCACCATGATAGCTCACCCAAAGCTCGACCCAGTTTCCGGTGAGCTATTTGCTCTTAGCTACGATGTGATTCAGAAGCCATACCTCAAGTACTTCAGATTTTCAAAAAATGGGGAAAAATCAAATGATGTTGAAATTCCAGTTGAAGACCCAACAATGATGCATGATTTCGCAATTACTGAGAACTTCGTCGTCATTCCTGATCAACAAGTCGTTTTCAAGATGTCTGAAATGATCCGTGGAGGTTCACCGGTGGTTTACGACAAGAACAAAGTTTCCCGATTTGGTATTCTGGATAAGTACGCGAAAGATGGGTCTGATTTGAAATGGGTTGAAGTACCTGATTGTTTCTGTTTCCACCTCTGGAATGCTTGGGAAGAAGCAGAAACAGATGAAATCGTTGTAATTGGTTCATGTATGACACCACCAGACTCCATTTTCAATGAATGTGATGAAGGGCTAAAGAGTGTTTTATCCGAAATCCGTCTCAATTTGAAAACAGGGAAATCAACAAGAAAATCCATAATCGAAAACCCGGATGAACAAGTGAATTTAGAAGCTGGAATGGTGAACCGAAACAAACTCGGAAGGAAAACAGAGTATGCTTATTTGGCTATCGCTGAACCATGGCCAAAAGTTTCTGGTTTTGCAAAAGTAAACCTGTTCACCGGTGAAGTTGAGAAATTCATTTATGGTGACAACAAATATGGTGGGGAACCTCTTTTTTTACCAAGAGACCCCAACAGCAAGGAAGAAGACGATGGTTATATTTTAGCTTTCGTTCACGATGAGAAAGAATGGAAATCAGAACTGCAAATTGTTAACGCAATGAGTTTGAAGTTGGAGGCAACTGTGAAGCTTCCATCAAGAGTTCCTTATGGATTTCATGGAACATTCATAAACGCCAATGATTTGGCAAATCAGGCATGA
| Download sequence region |
Get flanking sequences on SL2.50ch07
|
 mRNA Solyc07g056570.1.1 mRNA Solyc07g056570.1.1
 mRNA Solyc07g056570.1.1 mRNA Solyc07g056570.1.1
|
 Ontology terms Ontology terms
 Ontology terms Ontology terms
| terms associated with this mRNA |
 cDNA sequence cDNA sequence
 cDNA sequence cDNA sequence
| spliced cDNA sequence, including UTRs |
>Solyc07g056570.1.1 9-cis-epoxycarotenoid dioxygenase
ATGGCAACTACTACTTCACATGCCACAAATACATGGATTAAGACTAAGTTGTCAATGCCATCATCAAAGGAGTTTGGTTTTGCATCAAACTCTATTTCTCTACTCAAAAATCAACATAATAGGCAAAGTCTCAACATTAATTCCTCTCTTCAAGCTCCACCTATACTTCATTTTCCTAAACAATCTTCAAATTATCAAACACCAAAGAATAATACAATTTCACACCCAAAACAAGAAAACAACAACTCCTCTTCTTCTTCAACTTCCAAGTGGAATTTAGTGCAGAAAGCAGCAGCAATGGCTTTAGATGCTGTAGAAAGTGCTTTAACTAAACATGAACTTGAACACCCTTTGCCGAAAACAGCCGACCCACGAGTCCAGATTTCTGGGAATTTTGCTCCGGTACCGGAAAATCCAGTCTGTCAATCTCTTCCGGTCACCGGAAAAATACCCAAATGTGTTCAAGGCGTTTACGTTCGAAACGGAGCTAACCCTCTTTTTGAACCAACCGCCGGACACCATTTCTTCGACGGCGACGGTATGGTTCACGCCGTTCAATTCAAAAATGGGTCGGCTAGTTACGCTTGCCGTTTCACTGAAACAGAGAGGCTTGTTCAAGAAAAAGCTTTGGGTCGCCCTGTTTTCCCTAAAGCCATTGGTGAATTACATGGTCACTCTGGAATTGCAAGGCTTATGCTGTTTTACGCTCGTGGGCTCTTCGGACTTGTTGATCACAGTAAAGGAACTGGTGTTGCAAACGCCGGTTTAGTCTATTTCAATAACCGATTACTTGCTATGTCTGAAGATGATTTGCCTTACCATGTAAAGGTAACACCCACCGGCGATCTTAAAACAGAGGGTCGATTCGATTTCGACGGCCAGCTAAAATCCACCATGATAGCTCACCCAAAGCTCGACCCAGTTTCCGGTGAGCTATTTGCTCTTAGCTACGATGTGATTCAGAAGCCATACCTCAAGTACTTCAGATTTTCAAAAAATGGGGAAAAATCAAATGATGTTGAAATTCCAGTTGAAGACCCAACAATGATGCATGATTTCGCAATTACTGAGAACTTCGTCGTCATTCCTGATCAACAAGTCGTTTTCAAGATGTCTGAAATGATCCGTGGAGGTTCACCGGTGGTTTACGACAAGAACAAAGTTTCCCGATTTGGTATTCTGGATAAGTACGCGAAAGATGGGTCTGATTTGAAATGGGTTGAAGTACCTGATTGTTTCTGTTTCCACCTCTGGAATGCTTGGGAAGAAGCAGAAACAGATGAAATCGTTGTAATTGGTTCATGTATGACACCACCAGACTCCATTTTCAATGAATGTGATGAAGGGCTAAAGAGTGTTTTATCCGAAATCCGTCTCAATTTGAAAACAGGGAAATCAACAAGAAAATCCATAATCGAAAACCCGGATGAACAAGTGAATTTAGAAGCTGGAATGGTGAACCGAAACAAACTCGGAAGGAAAACAGAGTATGCTTATTTGGCTATCGCTGAACCATGGCCAAAAGTTTCTGGTTTTGCAAAAGTAAACCTGTTCACCGGTGAAGTTGAGAAATTCATTTATGGTGACAACAAATATGGTGGGGAACCTCTTTTTTTACCAAGAGACCCCAACAGCAAGGAAGAAGACGATGGTTATATTTTAGCTTTCGTTCACGATGAGAAAGAATGGAAATCAGAACTGCAAATTGTTAACGCAATGAGTTTGAAGTTGGAGGCAACTGTGAAGCTTCCATCAAGAGTTCCTTATGGATTTCATGGAACATTCATAAACGCCAATGATTTGGCAAATCAGGCATGA
ATGGCAACTACTACTTCACATGCCACAAATACATGGATTAAGACTAAGTTGTCAATGCCATCATCAAAGGAGTTTGGTTTTGCATCAAACTCTATTTCTCTACTCAAAAATCAACATAATAGGCAAAGTCTCAACATTAATTCCTCTCTTCAAGCTCCACCTATACTTCATTTTCCTAAACAATCTTCAAATTATCAAACACCAAAGAATAATACAATTTCACACCCAAAACAAGAAAACAACAACTCCTCTTCTTCTTCAACTTCCAAGTGGAATTTAGTGCAGAAAGCAGCAGCAATGGCTTTAGATGCTGTAGAAAGTGCTTTAACTAAACATGAACTTGAACACCCTTTGCCGAAAACAGCCGACCCACGAGTCCAGATTTCTGGGAATTTTGCTCCGGTACCGGAAAATCCAGTCTGTCAATCTCTTCCGGTCACCGGAAAAATACCCAAATGTGTTCAAGGCGTTTACGTTCGAAACGGAGCTAACCCTCTTTTTGAACCAACCGCCGGACACCATTTCTTCGACGGCGACGGTATGGTTCACGCCGTTCAATTCAAAAATGGGTCGGCTAGTTACGCTTGCCGTTTCACTGAAACAGAGAGGCTTGTTCAAGAAAAAGCTTTGGGTCGCCCTGTTTTCCCTAAAGCCATTGGTGAATTACATGGTCACTCTGGAATTGCAAGGCTTATGCTGTTTTACGCTCGTGGGCTCTTCGGACTTGTTGATCACAGTAAAGGAACTGGTGTTGCAAACGCCGGTTTAGTCTATTTCAATAACCGATTACTTGCTATGTCTGAAGATGATTTGCCTTACCATGTAAAGGTAACACCCACCGGCGATCTTAAAACAGAGGGTCGATTCGATTTCGACGGCCAGCTAAAATCCACCATGATAGCTCACCCAAAGCTCGACCCAGTTTCCGGTGAGCTATTTGCTCTTAGCTACGATGTGATTCAGAAGCCATACCTCAAGTACTTCAGATTTTCAAAAAATGGGGAAAAATCAAATGATGTTGAAATTCCAGTTGAAGACCCAACAATGATGCATGATTTCGCAATTACTGAGAACTTCGTCGTCATTCCTGATCAACAAGTCGTTTTCAAGATGTCTGAAATGATCCGTGGAGGTTCACCGGTGGTTTACGACAAGAACAAAGTTTCCCGATTTGGTATTCTGGATAAGTACGCGAAAGATGGGTCTGATTTGAAATGGGTTGAAGTACCTGATTGTTTCTGTTTCCACCTCTGGAATGCTTGGGAAGAAGCAGAAACAGATGAAATCGTTGTAATTGGTTCATGTATGACACCACCAGACTCCATTTTCAATGAATGTGATGAAGGGCTAAAGAGTGTTTTATCCGAAATCCGTCTCAATTTGAAAACAGGGAAATCAACAAGAAAATCCATAATCGAAAACCCGGATGAACAAGTGAATTTAGAAGCTGGAATGGTGAACCGAAACAAACTCGGAAGGAAAACAGAGTATGCTTATTTGGCTATCGCTGAACCATGGCCAAAAGTTTCTGGTTTTGCAAAAGTAAACCTGTTCACCGGTGAAGTTGAGAAATTCATTTATGGTGACAACAAATATGGTGGGGAACCTCTTTTTTTACCAAGAGACCCCAACAGCAAGGAAGAAGACGATGGTTATATTTTAGCTTTCGTTCACGATGAGAAAGAATGGAAATCAGAACTGCAAATTGTTAACGCAATGAGTTTGAAGTTGGAGGCAACTGTGAAGCTTCCATCAAGAGTTCCTTATGGATTTCATGGAACATTCATAAACGCCAATGATTTGGCAAATCAGGCATGA
 Protein sequence Protein sequence
 Protein sequence Protein sequence
| translated polypeptide sequence |
>Solyc07g056570.1.1 9-cis-epoxycarotenoid dioxygenase
MATTTSHATNTWIKTKLSMPSSKEFGFASNSISLLKNQHNRQSLNINSSLQAPPILHFPKQSSNYQTPKNNTISHPKQENNNSSSSSTSKWNLVQKAAAMALDAVESALTKHELEHPLPKTADPRVQISGNFAPVPENPVCQSLPVTGKIPKCVQGVYVRNGANPLFEPTAGHHFFDGDGMVHAVQFKNGSASYACRFTETERLVQEKALGRPVFPKAIGELHGHSGIARLMLFYARGLFGLVDHSKGTGVANAGLVYFNNRLLAMSEDDLPYHVKVTPTGDLKTEGRFDFDGQLKSTMIAHPKLDPVSGELFALSYDVIQKPYLKYFRFSKNGEKSNDVEIPVEDPTMMHDFAITENFVVIPDQQVVFKMSEMIRGGSPVVYDKNKVSRFGILDKYAKDGSDLKWVEVPDCFCFHLWNAWEEAETDEIVVIGSCMTPPDSIFNECDEGLKSVLSEIRLNLKTGKSTRKSIIENPDEQVNLEAGMVNRNKLGRKTEYAYLAIAEPWPKVSGFAKVNLFTGEVEKFIYGDNKYGGEPLFLPRDPNSKEEDDGYILAFVHDEKEWKSELQIVNAMSLKLEATVKLPSRVPYGFHGTFINANDLANQA*
MATTTSHATNTWIKTKLSMPSSKEFGFASNSISLLKNQHNRQSLNINSSLQAPPILHFPKQSSNYQTPKNNTISHPKQENNNSSSSSTSKWNLVQKAAAMALDAVESALTKHELEHPLPKTADPRVQISGNFAPVPENPVCQSLPVTGKIPKCVQGVYVRNGANPLFEPTAGHHFFDGDGMVHAVQFKNGSASYACRFTETERLVQEKALGRPVFPKAIGELHGHSGIARLMLFYARGLFGLVDHSKGTGVANAGLVYFNNRLLAMSEDDLPYHVKVTPTGDLKTEGRFDFDGQLKSTMIAHPKLDPVSGELFALSYDVIQKPYLKYFRFSKNGEKSNDVEIPVEDPTMMHDFAITENFVVIPDQQVVFKMSEMIRGGSPVVYDKNKVSRFGILDKYAKDGSDLKWVEVPDCFCFHLWNAWEEAETDEIVVIGSCMTPPDSIFNECDEGLKSVLSEIRLNLKTGKSTRKSIIENPDEQVNLEAGMVNRNKLGRKTEYAYLAIAEPWPKVSGFAKVNLFTGEVEKFIYGDNKYGGEPLFLPRDPNSKEEDDGYILAFVHDEKEWKSELQIVNAMSLKLEATVKLPSRVPYGFHGTFINANDLANQA*
 Gene model matches Gene model matches
 Gene model matches Gene model matches
|
 SGN Unigenes SGN Unigenes
 SGN Unigenes SGN Unigenes
| [Associate new unigene] |
Unigene ID:
[loading...]
 GenBank accessions GenBank accessions
 GenBank accessions GenBank accessions
| [Associate new genbank sequence] |
| Other genome matches | None |
 Literature annotations [9] Literature annotations [9]
 Literature annotations [9] Literature annotations [9]
| [Associate publication] [Matching publications] |
Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maize Vp14.
The Plant journal : for cell and molecular biology (1999)
Show / hide abstract
Show / hide abstract
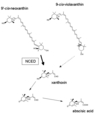
The notabilis (not) mutant of tomato has a wilty phenotype due to a deficiency in the levels of the plant hormone abscisic acid (ABA). The mutant appears to have a defect in a key control step in ABA biosynthesis--the oxidative cleavage of a 9-cis xanthophyll precursor to form the C15 intermediate, xanthoxin. A maize mutant, viviparous 14 (vp14) was recently obtained by transposon mutagenesis. This maize genetic lesion also affects the oxidative cleavage step in ABA synthesis. Degenerate primers for PCR, based on the VP14 predicted amino acid sequence, have been used to provide probes for screening a wilt-related tomato cDNA library. A full-length cDNA clone was identified which is specific to the not gene locus. The ORFs of the tomato cDNA and maize Vp14 are very similar, apart from parts of their N-terminal sequences. The not mutation has been characterized at the DNA level. A specific A/T base pair deletion of the coding sequence has resulted in a frameshift mutation, indicating that not is a null mutant. This observation is discussed in connection with the relatively mild phenotype exhibited by not mutant homozygotes.
Burbidge, A. Grieve, T. Jackson, A. Thompson, A. McCarty, D. Taylor, I.
The Plant journal : for cell and molecular biology.
1999.
17(4).
427-31.
Abscisic acid biosynthesis in tomato: regulation of zeaxanthin epoxidase and 9-cis-epoxycarotenoid dioxygenase mRNAs by light/dark cycles, water stress and abscisic acid.
Plant molecular biology (2000)
Show / hide abstract
Show / hide abstract
Two genes encoding enzymes in the abscisic acid (ABA) biosynthesis pathway, zeaxanthin epoxidase (ZEP) and 9-cis-epoxycarotenoid dioxygenase (NCED), have previously been cloned by transposon tagging in Nicotiana plumbaginifolia and maize respectively. We demonstrate that antisense down-regulation of the tomato gene LeZEP1 causes accumulation of zeaxanthin in leaves, suggesting that this gene also encodes ZEP. LeNCED1 is known to encode NCED from characterization of a null mutation (notabilis) in tomato. We have used LeZEP1 and LeNCED1 as probes to study gene expression in leaves and roots of whole plants given drought treatments, during light/dark cycles, and during dehydration of detached leaves. During drought stress, NCED mRNA increased in both leaves and roots, whereas ZEP mRNA increased in roots but not leaves. When detached leaves were dehydrated, NCED mRNA responded rapidly to small reductions in water content. Using a detached leaf system with ABA-deficient mutants and ABA feeding, we investigated the possibility that NCED mRNA is regulated by the end product of the pathway, ABA, but found no evidence that this is the case. We also describe strong diurnal expression patterns for both ZEP and NCED, with the two genes displaying distinctly different patterns. ZEP mRNA oscillated with a phase very similar to light-harvesting complex II (LHCII) mRNA, and oscillations continued in a 48 h dark period. NCED mRNA oscillated with a different phase and remained low during a 48 h dark period. Implications for regulation of water stress-induced ABA biosynthesis are discussed.
Thompson, A. Jackson, A. Parker, R. Morpeth, D. Burbidge, A. Taylor, I.
Plant molecular biology.
2000.
42(6).
833-45.
Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid.
The Plant journal : for cell and molecular biology (2000)
Show / hide abstract
Show / hide abstract
The tomato mutant notabilis has a wilty phenotype as a result of abscisic acid (ABA) deficiency. The wild-type allele of notabilis, LeNCED1, encodes a putative 9-cis-epoxycarotenoid dioxygenase (NCED) with a potential regulatory role in ABA biosynthesis. We have created transgenic tobacco plants in which expression of the LeNCED1 coding region is under tetracycline-inducible control. When leaf explants from these plants were treated with tetracycline, NCED mRNA was induced and bulk leaf ABA content increased by up to 10-fold. Transgenic tomato plants were also produced containing the LeNCED1 coding region under the control of one of two strong constitutive promoters, either the doubly enhanced CaMV 35S promoter or the chimaeric 'Super-Promoter'. Many of these plants were wilty, suggesting co-suppression of endogenous gene activity; however three transformants displayed a common, heritable phenotype that could be due to enhanced ABA biosynthesis, showing increased guttation and seed dormancy. Progeny from two of these transformants were further characterized, and it was shown that they also exhibited reduced stomatal conductance, increased NCED mRNA and elevated seed ABA content. Progeny of one transformant had significantly higher bulk leaf ABA content compared to the wild type. The increased seed dormancy was reversed by addition of the carotenoid biosynthesis inhibitor norflurazon. These data provide strong evidence that NCED is indeed a key regulatory enzyme in ABA biosynthesis in leaves, and demonstrate for the first time that plant ABA content can be increased through manipulating NCED.
Thompson, A. Jackson, A. Symonds, R. Mulholland, B. Dadswell, A. Blake, P. Burbidge, A. Taylor, I.
The Plant journal : for cell and molecular biology.
2000.
23(3).
363-74.
Abnormal Stomatal Behavior in Wilty Mutants of Tomato.
(1966)
Show / hide abstract
Show / hide abstract
An attempt was made to explain the excessive wilting tendency of 3 tomato mutants, notabilis, flacca, and sitiens. The control varieties in which these mutations were induced are Rheinlands Ruhm for flacca and sitiens and Lukullus for notabilis. Although all 3 mutants are alleles of separated loci, they seem to react similarly to water stress. The mutants wilt faster than the control plants when both are subjected to the same water stress. It was demonstrated by measurements of water loss from whole plants that all 3 mutants have much higher rates of transpiration than the control varieties, particularly at night. The extent of cuticular transpiration was compared in both kinds of plants by measuring the rate of water loss from detached drying leaves coated with vaseline on the lower surface. The difference in cuticular transpiration between the mutant and the control plants seems to be negligible. However, various facts point to stomata as the main factor responsible for the higher rates of water loss in the mutant plants. The stomata of the latter tend to open wider and to resist closure in darkness, in wilted leaves, and when treated with phenylmercuric acetate. Stomata of the 2 extreme mutants, sitiens and flacca, remain open even when the guard cells are plasmolyzed. The stomata of the mutants also are more frequent per unit of leaf surface and vary more in their size.
Tal, Moshe.
.
1966.
41(8).
1387-1391.
Regulation and manipulation of ABA biosynthesis in roots.
Plant, cell & environment (2007)
Show / hide abstract
Show / hide abstract
Overexpression of 9-cis-epoxycarotenoid dioxygenase (NCED) is known to cause abscisic acid (ABA) accumulation in leaves, seeds and whole plants. Here we investigated the manipulation of ABA biosynthesis in roots. Roots from whole tomato plants that constitutively overexpress LeNCED1 had a higher ABA content than wild-type (WT) roots. This could be explained by enhanced in situ ABA biosynthesis, rather than import of ABA from the shoot, because root cultures also had higher ABA content, and because tetracycline (Tc)-induced LeNCED1 expression caused ABA accumulation in isolated tobacco roots. However, the Tc-induced expression led to greater accumulation of ABA in leaves than in roots. This demonstrates for the first time that NCED is rate-limiting in root tissues, but suggests that other steps were also restrictive to pathway flux, more so in roots than in leaves. Dehydration and NCED overexpression acted synergistically in enhancing ABA accumulation in tomato root cultures. One explanation is that xanthophyll synthesis was increased during root dehydration, and, in support of this, dehydration treatments increased beta-carotene hydroxylase mRNA levels. Whole plants overexpressing LeNCED1 exhibited greatly reduced stomatal conductance and grafting experiments from this study demonstrated that this was predominantly due to increased ABA biosynthesis in leaves rather than in roots. Genetic manipulation of both xanthophyll supply and epoxycarotenoid cleavage may be needed to enhance root ABA biosynthesis sufficiently to signal stomatal closure in the shoot.
Thompson, Andrew. Mulholland, Barry. Jackson, Alison. McKee, John. Hilton, Howard. Symonds, Rachael. Sonneveld, Tineke. Burbidge, Alan. Stevenson, Patrick. Taylor, Ian.
Plant, cell & environment.
2007.
30(1).
67-78.
Overproduction of abscisic acid in tomato increases transpiration efficiency and root hydraulic conductivity and influences leaf expansion.
Plant physiology (2007)
Show / hide abstract
Show / hide abstract
Overexpression of genes that respond to drought stress is a seemingly attractive approach for improving drought resistance in crops. However, the consequences for both water-use efficiency and productivity must be considered if agronomic utility is sought. Here, we characterize two tomato (Solanum lycopersicum) lines (sp12 and sp5) that overexpress a gene encoding 9-cis-epoxycarotenoid dioxygenase, the enzyme that catalyzes a key rate-limiting step in abscisic acid (ABA) biosynthesis. Both lines contained more ABA than the wild type, with sp5 accumulating more than sp12. Both had higher transpiration efficiency because of their lower stomatal conductance, as demonstrated by increases in delta(13)C and delta(18)O, and also by gravimetric and gas-exchange methods. They also had greater root hydraulic conductivity. Under well-watered glasshouse conditions, mature sp5 plants were found to have a shoot biomass equal to the wild type despite their lower assimilation rate per unit leaf area. These plants also had longer petioles, larger leaf area, increased specific leaf area, and reduced leaf epinasty. When exposed to root-zone water deficits, line sp12 showed an increase in xylem ABA concentration and a reduction in stomatal conductance to the same final levels as the wild type, but from a different basal level. Indeed, the main difference between the high ABA plants and the wild type was their performance under well-watered conditions: the former conserved soil water by limiting maximum stomatal conductance per unit leaf area, but also, at least in the case of sp5, developed a canopy more suited to light interception, maximizing assimilation per plant, possibly due to improved turgor or suppression of epinasty.
Thompson, Andrew. Andrews, John. Mulholland, Barry. McKee, John. Hilton, Howard. Horridge, Jon. Farquhar, Graham. Smeeton, Rachel. Smillie, Ian. Black, Colin. Taylor, Ian.
Plant physiology.
2007.
143(4).
1905-17.
The role of ABA in triggering ethylene biosynthesis and ripening of tomato fruit.
Journal of experimental botany (2009)
Show / hide abstract
Show / hide abstract
In order to understand more details about the role of abscisic acid (ABA) in fruit ripening and senescence of tomato, two cDNAs (LeNCED1 and LeNCED2) which encode 9-cis-epoxycarotenoid dioxygenase (NCED) as a key enzyme in ABA biosynthesis, two cDNAs (LeACS2 and LeACS4) which encode 1-aminocyclopropane-1-carboxylic acid (ACC) synthase, and one cDNA (LeACO1) which encodes ACC oxidase involved in ethylene biosynthesis were cloned from tomato fruit using a reverse transcription-PCR (RT-PCR) approach. The relationship between ABA and ethylene during ripening was also investigated. Among six sampling times in tomato fruits, the LeNCED1 gene was highly expressed only at the breaker stage when the ABA content becomes high. After this, the LeACS2, LeACS4, and LeACO1 genes were expressed with some delay. The change in pattern of ACO activity was in accordance with ethylene production reaching its peak at the pink stage. The maximum ABA content preceded ethylene production in both the seeds and the flesh. The peak value of ABA, ACC, and ACC oxidase activity, and ethylene production all started to increase earlier in seeds than in flesh tissues, although they occurred at different ripening stages. Exogenous ABA treatment increased the ABA content in both flesh and seed, inducing the expression of both ACS and ACO genes, and promoting ethylene synthesis and fruit ripening, while treatment with fluridone or nordihydroguaiaretic acid (NDGA) inhibited them, delaying fruit ripening and softening. Based on the results obtained in this study, it was concluded that LeNCED1 initiates ABA biosynthesis at the onset of fruit ripening, and might act as an original inducer, and ABA accumulation might play a key role in the regulation of ripeness and senescence of tomato fruit.
Zhang, M. Yuan, B. Leng, P.
Journal of experimental botany.
2009.
60(6).
1579-88.
Ripening-associated ethylene biosynthesis in tomato fruit is autocatalytically and developmentally regulated.
Journal of experimental botany (2009)
Show / hide abstract
Show / hide abstract
To investigate the regulatory mechanism(s) of ethylene biosynthesis in fruit, transgenic tomatoes with all known LeEIL genes suppressed were produced by RNA interference engineering. The transgenic tomato exhibited ethylene insensitivity phenotypes such as non-ripening and the lack of the triple response and petiole epinasty of seedlings even in the presence of exogenous ethylene. Transgenic fruit exhibited a low but consistent increase in ethylene production beyond 40 days after anthesis (DAA), with limited LeACS2 and LeACS4 expression. 1-Methylcyclopropene (1-MCP), a potent inhibitor of ethylene perception, failed to inhibit the limited increase in ethylene production and expression of the two 1-aminocyclopropane-1-carboxylic acid (ACC) synthase (ACS) genes in the transgenic fruit. These results suggest that ripening-associated ethylene (system 2) in wild-type tomato fruit consists of two parts: a small part regulated by a developmental factor through the ethylene-independent expression of LeACS2 and LeACS4 and a large part regulated by an autocatalytic system due to the ethylene-dependent expression of the same genes. The results further suggest that basal ethylene (system 1) is less likely to be involved in the transition to system 2. Even if the effect of system 1 ethylene is eliminated, fruit can show a small increase in ethylene production due to unknown developmental factors. This increase would be enough for the stimulation of autocatalytic ethylene production, leading to fruit ripening.
Yokotani, N. Nakano, R. Imanishi, S. Nagata, M. Inaba, A. Kubo, Y.
Journal of experimental botany.
2009.
().
.
Fruit-specific RNAi-mediated suppression of SlNCED1 increases both lycopene and β-carotene contents in tomato fruit.
Journal of experimental botany (2012)
Show / hide abstract
Show / hide abstract
Abscisic acid (ABA) plays important roles during tomato fruit ripening. To study the regulation of carotenoid biosynthesis by ABA, the SlNCED1 gene encoding 9-cis-epoxycarotenoid dioxygenase (NCED), a key enzyme in the ABA biosynthesis, was suppressed in tomato plants by transformation with an RNA interference (RNAi) construct driven by a fruit-specific E8 promoter. ABA accumulation and SlNCED1 transcript levels in the transgenic fruit were down-regulated to between 20-50% of that in control fruit. This significant reduction in NCED activity led to the carbon that normally channels to free ABA as well as the ABA metabolite accumulation during ripening to be partially blocked. Therefore, this 'backlogged' carbon transformed into the carotenoid pathway in the RNAi lines resulted in increased assimilation and accumulation of upstream compounds in the pathway, chiefly lycopene and β-carotene. Fruit of all RNAi lines displayed deep red coloration compared with the pink colour of control fruit. The decrease in endogenous ABA in these transgenics resulted in an increase in ethylene, by increasing the transcription of genes related to the synthesis of ethylene during ripening. In conclusion, ABA potentially regulated the degree of pigmentation and carotenoid composition during ripening and could control, at least in part, ethylene production and action in climacteric tomato fruit.
Sun, L. Yuan, B. Zhang, M. Wang, L. Cui, M. Wang, Q. Leng, P.
Journal of experimental botany.
2012.
63(8).
3097-108.
 Ontology annotations (16) Ontology annotations (16)
 Ontology annotations (16) Ontology annotations (16)
| [Add ontology annotations] |
[loading...]
 Related views Related views
 Related views Related views
|
- Genomic details
| User comments |
Please wait, checking for comments. (If comments do not show up, access them here)
Your Lists
Public Lists
List Contents
List Validation Report: Failed
Elements not found:
Optional: Add Missing Accessions to A List
Mismatched case
Click the Adjust Case button to align the case in the list with what is in the database.
Multiple mismatched case
Items listed here have mulitple case mismatches and must be fixed manually. If accessions need to be merged, contact the database directly.
List elements matching a synonym
Multiple synonym matches
Fuzzy Search Results
Synonym Search Results
Available Seedlots
Your Datasets
Public Datasets
Dataset Contents
Dataset Validation Failed
Elements not found:
Your Calendar
Having trouble viewing events on the calendar?
Are you associated with the breeding program you are interested in viewing?
Add New Event
Event Info
| Attribute | Value |
|---|---|
| Project Name: | |
| Start Date: | |
| End Date: | |
| Event Type: | |
| Event Description: | |
| Event Web URL: |
Edit Event
Login
Forgot Username
If you've forgotten your username, enter your email address below. An email will be sent with any account username(s) associated with your email address.
Reset Password
To reset your password, please enter your email address. A link will be sent to that address with a link that will enable you to reset your password.
Create New User
Working