This site uses cookies to provide logins and other features. Please accept the use of cookies by clicking Accept.
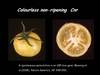
Tomato locus Colorless non-ripening
| Locus details | Download GMOD XML | Note to Editors | Annotation guidelines |
[loading edit links...]
|
[loading...]
|
|
 Links to external databases Links to external databases
 Links to external databases Links to external databases
|
| Registry name: | None | [Associate registry name] |
 Notes and figures (1) Notes and figures (1)
 Notes and figures (1) Notes and figures (1)
| [Add notes, figures or images] |
 Accessions and images (0) Accessions and images (0)
 Accessions and images (0) Accessions and images (0)
| [Associate accession] |
Accession name:
Would you Like to specify an allele?
| Alleles (0) | None | [Add new Allele] |
 Associated loci (0) Associated loci (0)
 Associated loci (0) Associated loci (0)
| [Associate new locus] |
[loading...]
|
| Associated loci - graphical view | None |
 SolCyc links SolCyc links
 SolCyc links SolCyc links
|
[loading...]
 Sequence annotations Sequence annotations
 Sequence annotations Sequence annotations
|
 Genome features Genome features
 Genome features Genome features
|
 Genomic sequence Genomic sequence
 Genomic sequence Genomic sequence
| unprocessed genomic sequence region underlying this gene |
>Solyc02g077920.2 SL2.50ch02:42745257..42743081 (sequence from reverse strand)
CCTTGACAAAAAGTTAGTGGAGTAACTACCTAGGAGTAAATTCAATAGTAGACCTTGAAAAGAACTTTAGCAAAGTCATCATAAATGCTCTTCACGTCTCATGTACTATGTTAAGGAATGGTCACATTTCTCTCTGCATTAAAGCTAGTTCATGTTAAAAGTTGAGGCCGGTAGTAGTTTCAACTTTCAATTTAATTCCACCTTTCCTGGCCCACTTCTGTACGGAACACCAATCAGAATCTTTAGTTCATCTTAACACCAAAGCATCTCCACTTAGACACTTACTAGACTTCACATAGGAGGAAAAATATGGAACTGGTGGTCCTCACACGTACTTACCTTTCTTTTTTTACCTTTGTTCAAGTTTCATACTCTTTTATCTGGCTTCCTCACTCTATTTTGGCCCAATAGGTTCTCCTCACAGGGATGGAAACTAACAAATGGGAAGGGAAGAGAAGCATTACTGAAGCTGAAAAGGAAGAGGATGAACATGGAAGTGTTGAAGAGGATAGCAAAAGAAAAAGGGTATTGACTCTCTCTGGTAGGAAGCTAGTTGGTGAAGGGTCGGCACATCCTTCTTGCCAGGTCGATCAGTGCACTGCAGATATGGCAGATGCCAAGCCATACCATCGCCGCCACAAGGTGTGTGAGTTCCATTCAAAGTCTCCAATAGTACTTATTAGTGGACTCCAGAAGCGATTCTGTCAGCAATGTAGCAGGTAAGTAACCCCTAATTTCTGATGATTATCCGTGCCTTCCGAGTTGTATAGTGGAGATATCTTGGAAACTAATTATGTGCACTCATCAAACAGTTTTTCTTAGATTTAAGTCATGAGGAACATTAATCAAAACATCAATTAGCCAATAAACAAAGAAGAAGGTAATTTAAGCTTGAGTGTTGATTCATTTTGTGCAACATATGGGAATCCTAATATCAAGAACCAATACCAATAACACAGGCAAAAACTAATACATAGAGACACGAAAGTACAACAGCAACAATCAATTTTATACAATATCCTATGCATATCATTTATTTTATCATAAACTGTGAAAAAGCAAAAGTGAGTCTTTTAAATCGTGATATAACAATGAATCCAAGACACTACTGGATCTTTTAATTAATCCATTGACATTTCTAGACTATTAACTTTGAAATGTGGTTATTAAGATTATGGCTACATCCAATTAAGAGATAGTTGTTAACCTCTAGGTTTCCTTGTTATGGAACAAAAGCCAAATCAAGCAATGATGATGACTTCTTAGAATTAATCTCACTGTCCAATGGGTCTTCAAGTTGACCAAAAAAATGTGAATAAATATTGCTAACATGGAGAATGGTCTGTATGGTTGCGATAGGGCTTCTTGATCGTCCCTTCTTTAGGTAGTAGAAGGTAAGCTTTACTCCAACTGAAATTGTATACAAAAGATTCTTTTGTTTTCTTTGAAGGGAGAGACAAATACTGGTAAGGGAAAGAATTAACTTTTGCACATGCAAACAGAGATTATCCTTCAGACAAATTTTTTGTTCCAAATTCATCCATCTGACAACGAGGTATTCTAAATAAGATTCTACTTCCTATGAAAGTATGTAGTGTATCTAATCAGCCAAGAAACTGTAGCTGCTGCCACATCCACAGCCATCTCATGTATTTTCTAATCAGCTGCTTTCAGATTCCCAAGTATAGGAGTATATCATTTTCTAATGAGCAGGGGCAAAACAGAGCTTTTAATGTGAATTTTCAATTGTTCGCATGATGAGGTGCAGATTTCATCTGTTAGCAGAGTTTGATGATGCTAAGAGGAGTTGCCGAAGGCGTTTGGCAGGTCACAATGAGCGCCGCCGTAAAATTACATATGACTCTCATGGAGAAAATTTGGGCTGAAGAAGCATCAGCATCAATGGCAGCCAAATAACCTACTTTCTGAAGCAACATAAACAAGATGTGGGTTAAGCATGCTCTCTATCTTCTGTCAATTCCCGGATTTCTAAGCAAATTGTTATTTGTGACCTTCAAGTATCTAGCTAATAGTACTTTCATTTCCCTTGTGTAAAGGGAAGGACAAAAAGAATAAGCCGTTGGCACTGTCTGCTAGTATTCGAAAAATGAAATATTTGTAGTATGAATGCATATCCATGTTGACATTCCAACTCACTTTATGAGGTTTTTATTATT
CCTTGACAAAAAGTTAGTGGAGTAACTACCTAGGAGTAAATTCAATAGTAGACCTTGAAAAGAACTTTAGCAAAGTCATCATAAATGCTCTTCACGTCTCATGTACTATGTTAAGGAATGGTCACATTTCTCTCTGCATTAAAGCTAGTTCATGTTAAAAGTTGAGGCCGGTAGTAGTTTCAACTTTCAATTTAATTCCACCTTTCCTGGCCCACTTCTGTACGGAACACCAATCAGAATCTTTAGTTCATCTTAACACCAAAGCATCTCCACTTAGACACTTACTAGACTTCACATAGGAGGAAAAATATGGAACTGGTGGTCCTCACACGTACTTACCTTTCTTTTTTTACCTTTGTTCAAGTTTCATACTCTTTTATCTGGCTTCCTCACTCTATTTTGGCCCAATAGGTTCTCCTCACAGGGATGGAAACTAACAAATGGGAAGGGAAGAGAAGCATTACTGAAGCTGAAAAGGAAGAGGATGAACATGGAAGTGTTGAAGAGGATAGCAAAAGAAAAAGGGTATTGACTCTCTCTGGTAGGAAGCTAGTTGGTGAAGGGTCGGCACATCCTTCTTGCCAGGTCGATCAGTGCACTGCAGATATGGCAGATGCCAAGCCATACCATCGCCGCCACAAGGTGTGTGAGTTCCATTCAAAGTCTCCAATAGTACTTATTAGTGGACTCCAGAAGCGATTCTGTCAGCAATGTAGCAGGTAAGTAACCCCTAATTTCTGATGATTATCCGTGCCTTCCGAGTTGTATAGTGGAGATATCTTGGAAACTAATTATGTGCACTCATCAAACAGTTTTTCTTAGATTTAAGTCATGAGGAACATTAATCAAAACATCAATTAGCCAATAAACAAAGAAGAAGGTAATTTAAGCTTGAGTGTTGATTCATTTTGTGCAACATATGGGAATCCTAATATCAAGAACCAATACCAATAACACAGGCAAAAACTAATACATAGAGACACGAAAGTACAACAGCAACAATCAATTTTATACAATATCCTATGCATATCATTTATTTTATCATAAACTGTGAAAAAGCAAAAGTGAGTCTTTTAAATCGTGATATAACAATGAATCCAAGACACTACTGGATCTTTTAATTAATCCATTGACATTTCTAGACTATTAACTTTGAAATGTGGTTATTAAGATTATGGCTACATCCAATTAAGAGATAGTTGTTAACCTCTAGGTTTCCTTGTTATGGAACAAAAGCCAAATCAAGCAATGATGATGACTTCTTAGAATTAATCTCACTGTCCAATGGGTCTTCAAGTTGACCAAAAAAATGTGAATAAATATTGCTAACATGGAGAATGGTCTGTATGGTTGCGATAGGGCTTCTTGATCGTCCCTTCTTTAGGTAGTAGAAGGTAAGCTTTACTCCAACTGAAATTGTATACAAAAGATTCTTTTGTTTTCTTTGAAGGGAGAGACAAATACTGGTAAGGGAAAGAATTAACTTTTGCACATGCAAACAGAGATTATCCTTCAGACAAATTTTTTGTTCCAAATTCATCCATCTGACAACGAGGTATTCTAAATAAGATTCTACTTCCTATGAAAGTATGTAGTGTATCTAATCAGCCAAGAAACTGTAGCTGCTGCCACATCCACAGCCATCTCATGTATTTTCTAATCAGCTGCTTTCAGATTCCCAAGTATAGGAGTATATCATTTTCTAATGAGCAGGGGCAAAACAGAGCTTTTAATGTGAATTTTCAATTGTTCGCATGATGAGGTGCAGATTTCATCTGTTAGCAGAGTTTGATGATGCTAAGAGGAGTTGCCGAAGGCGTTTGGCAGGTCACAATGAGCGCCGCCGTAAAATTACATATGACTCTCATGGAGAAAATTTGGGCTGAAGAAGCATCAGCATCAATGGCAGCCAAATAACCTACTTTCTGAAGCAACATAAACAAGATGTGGGTTAAGCATGCTCTCTATCTTCTGTCAATTCCCGGATTTCTAAGCAAATTGTTATTTGTGACCTTCAAGTATCTAGCTAATAGTACTTTCATTTCCCTTGTGTAAAGGGAAGGACAAAAAGAATAAGCCGTTGGCACTGTCTGCTAGTATTCGAAAAATGAAATATTTGTAGTATGAATGCATATCCATGTTGACATTCCAACTCACTTTATGAGGTTTTTATTATT
| Download sequence region |
Get flanking sequences on SL2.50ch02
|
 mRNA Solyc02g077920.2.1 mRNA Solyc02g077920.2.1
 mRNA Solyc02g077920.2.1 mRNA Solyc02g077920.2.1
|
 Ontology terms Ontology terms
 Ontology terms Ontology terms
| terms associated with this mRNA |
 cDNA sequence cDNA sequence
 cDNA sequence cDNA sequence
| spliced cDNA sequence, including UTRs |
>Solyc02g077920.2.1 Squamosa promoter binding-like protein (AHRD V1 ***- Q0PY35_SOLLC); contains Interpro domain(s) IPR004333 Transcription factor, SBP-box
CCTTGACAAAAAGTTAGTGGAGTAACTACCTAGGAGTAAATTCAATAGTAGACCTTGAAAAGAACTTTAGCAAAGTCATCATAAATGCTCTTCACGTCTCATGTACTATGTTAAGGAATGGTCACATTTCTCTCTGCATTAAAGCTAGTTCATGTTAAAAGTTGAGGCCGGTAGTAGTTTCAACTTTCAATTTAATTCCACCTTTCCTGGCCCACTTCTGTACGGAACACCAATCAGAATCTTTAGTTCATCTTAACACCAAAGCATCTCCACTTAGACACTTACTAGACTTCACATAGGAGGAAAAATATGGAACTGGTGGTCCTCACACGTTCTCCTCACAGGGATGGAAACTAACAAATGGGAAGGGAAGAGAAGCATTACTGAAGCTGAAAAGGAAGAGGATGAACATGGAAGTGTTGAAGAGGATAGCAAAAGAAAAAGGGTATTGACTCTCTCTGGTAGGAAGCTAGTTGGTGAAGGGTCGGCACATCCTTCTTGCCAGGTCGATCAGTGCACTGCAGATATGGCAGATGCCAAGCCATACCATCGCCGCCACAAGGTGTGTGAGTTCCATTCAAAGTCTCCAATAGTACTTATTAGTGGACTCCAGAAGCGATTCTGTCAGCAATGTAGCAGATTTCATCTGTTAGCAGAGTTTGATGATGCTAAGAGGAGTTGCCGAAGGCGTTTGGCAGGTCACAATGAGCGCCGCCGTAAAATTACATATGACTCTCATGGAGAAAATTTGGGCTGAAGAAGCATCAGCATCAATGGCAGCCAAATAACCTACTTTCTGAAGCAACATAAACAAGATGTGGGTTAAGCATGCTCTCTATCTTCTGTCAATTCCCGGATTTCTAAGCAAATTGTTATTTGTGACCTTCAAGTATCTAGCTAATAGTACTTTCATTTCCCTTGTGTAAAGGGAAGGACAAAAAGAATAAGCCGTTGGCACTGTCTGCTAGTATTCGAAAAATGAAATATTTGTAGTATGAATGCATATCCATGTTGACATTCCAACTCACTTTATGAGGTTTTTATTATT
CCTTGACAAAAAGTTAGTGGAGTAACTACCTAGGAGTAAATTCAATAGTAGACCTTGAAAAGAACTTTAGCAAAGTCATCATAAATGCTCTTCACGTCTCATGTACTATGTTAAGGAATGGTCACATTTCTCTCTGCATTAAAGCTAGTTCATGTTAAAAGTTGAGGCCGGTAGTAGTTTCAACTTTCAATTTAATTCCACCTTTCCTGGCCCACTTCTGTACGGAACACCAATCAGAATCTTTAGTTCATCTTAACACCAAAGCATCTCCACTTAGACACTTACTAGACTTCACATAGGAGGAAAAATATGGAACTGGTGGTCCTCACACGTTCTCCTCACAGGGATGGAAACTAACAAATGGGAAGGGAAGAGAAGCATTACTGAAGCTGAAAAGGAAGAGGATGAACATGGAAGTGTTGAAGAGGATAGCAAAAGAAAAAGGGTATTGACTCTCTCTGGTAGGAAGCTAGTTGGTGAAGGGTCGGCACATCCTTCTTGCCAGGTCGATCAGTGCACTGCAGATATGGCAGATGCCAAGCCATACCATCGCCGCCACAAGGTGTGTGAGTTCCATTCAAAGTCTCCAATAGTACTTATTAGTGGACTCCAGAAGCGATTCTGTCAGCAATGTAGCAGATTTCATCTGTTAGCAGAGTTTGATGATGCTAAGAGGAGTTGCCGAAGGCGTTTGGCAGGTCACAATGAGCGCCGCCGTAAAATTACATATGACTCTCATGGAGAAAATTTGGGCTGAAGAAGCATCAGCATCAATGGCAGCCAAATAACCTACTTTCTGAAGCAACATAAACAAGATGTGGGTTAAGCATGCTCTCTATCTTCTGTCAATTCCCGGATTTCTAAGCAAATTGTTATTTGTGACCTTCAAGTATCTAGCTAATAGTACTTTCATTTCCCTTGTGTAAAGGGAAGGACAAAAAGAATAAGCCGTTGGCACTGTCTGCTAGTATTCGAAAAATGAAATATTTGTAGTATGAATGCATATCCATGTTGACATTCCAACTCACTTTATGAGGTTTTTATTATT
 Protein sequence Protein sequence
 Protein sequence Protein sequence
| translated polypeptide sequence |
>Solyc02g077920.2.1 Squamosa promoter binding-like protein (AHRD V1 ***- Q0PY35_SOLLC); contains Interpro domain(s) IPR004333 Transcription factor, SBP-box
METNKWEGKRSITEAEKEEDEHGSVEEDSKRKRVLTLSGRKLVGEGSAHPSCQVDQCTADMADAKPYHRRHKVCEFHSKSPIVLISGLQKRFCQQCSRFHLLAEFDDAKRSCRRRLAGHNERRRKITYDSHGENLG*
METNKWEGKRSITEAEKEEDEHGSVEEDSKRKRVLTLSGRKLVGEGSAHPSCQVDQCTADMADAKPYHRRHKVCEFHSKSPIVLISGLQKRFCQQCSRFHLLAEFDDAKRSCRRRLAGHNERRRKITYDSHGENLG*
 Gene model matches Gene model matches
 Gene model matches Gene model matches
|
 SGN Unigenes SGN Unigenes
 SGN Unigenes SGN Unigenes
| [Associate new unigene] |
Unigene ID:
[loading...]
 GenBank accessions GenBank accessions
 GenBank accessions GenBank accessions
| [Associate new genbank sequence] |
DQ672601 Lycopersicon esculentum cultivar Ailsa Craig leucine-rich repeat family protein, putative polyprotein, auxin repressed/dormancy associated protein, putative zinc finger protein, putative protease/hydrolase, mitochondrial amino acid carrier, squamosa promoter binding-like protein, putative retroelement pol polyprotein, and Dof zinc finger protein genes, complete cds.
| Other genome matches | None |
 Literature annotations [7] Literature annotations [7]
 Literature annotations [7] Literature annotations [7]
| [Associate publication] [Matching publications] |
Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant
(1999)
Show / hide abstract
Show / hide abstract
In this paper we describe a novel, dominant pleiotropic tomato (Lycopersicon esculentum)-ripening mutation, Cnr (colorless nonripening). This mutant occurred spontaneously in a commercial population. Cnr has a phenotype that is quite distinct from that of the other pleiotropic tomato-ripening mutants and is characterized by fruit that show greatly reduced ethylene production, an inhibition of softening, a yellow skin, and a nonpigmented pericarp. The ripening-related biosynthesis of carotenoid pigments was abolished in the pericarp tissue. The pericarp also showed a significant reduction in cell-to-cell adhesion, with cell separation occurring when blocks of tissue were incubated in water alone. The mutant phenotype was not reversed by exposure to exogenous ethylene. Crosses with other mutant lines and the use of a restriction fragment length polymorphism marker demonstrated that Cnr was not allelic with the pleiotropic ripening mutants nor, alc, rin, Nr, Gr, and Nr-2. The gene has been mapped to the top of chromosome 2, also indicating that it is distinct from the other pleiotropic ripening mutants. We undertook the molecular characterization of Cnr by examining the expression of a panel of ripening-related genes in the presence and absence of exogenous ethylene. The pattern of gene expression in Cnr was related to, but differed from, that of several of the other well-characterized mutants. We discuss here the possible relationships among nor, Cnr, and rin in a putative ripening signal cascade.
Thompson, AJ. Tor, M. Barry, CS. Vrebalov, J. Orfila, C. Jarvis, MC. Giovannoni, JJ. Grierson, D. Seymour, GB.
.
1999.
120(2).
383-90.
Altered middle lamella homogalacturonan and disrupted deposition of (1-->5)-alpha-L-arabinan in the pericarp of Cnr, a ripening mutant of tomato.
Plant physiology (2001)
Show / hide abstract
Show / hide abstract
Cnr (colorless non-ripening) is a pleiotropic tomato (Lycopersicon esculentum) fruit ripening mutant with altered tissue properties including weaker cell-to-cell contacts in the pericarp (A.J. Thompson, M. Tor, C.S. Barry, J. Vrebalov, C. Orfila, M.C. Jarvis, J.J. Giovannoni, D. Grierson, G.B. Seymour [1999] Plant Physiol 120: 383-390). Whereas the genetic basis of the Cnr mutation is being identified by molecular analyses, here we report the identification of cell biological factors underlying the Cnr texture phenotype. In comparison with wild type, ripe-stage Cnr fruits have stronger, non-swollen cell walls (CW) throughout the pericarp and extensive intercellular space in the inner pericarp. Using electron energy loss spectroscopy imaging of calcium-binding capacity and anti-homogalacturonan (HG) antibody probes (PAM1 and JIM5) we demonstrate that maturation processes involving middle lamella HG are altered in Cnr fruit, resulting in the absence or a low level of HG-/calcium-based cell adhesion. We also demonstrate that the deposition of (1-->5)-alpha-L-arabinan is disrupted in Cnr pericarp CW and that this disruption occurs prior to fruit ripening. The relationship between the disruption of (1-->5)-alpha-L-arabinan deposition in pericarp CW and the Cnr phenotype is discussed.
Orfila, C. Seymour, G. Willats, W. Huxham, I. Jarvis, M. Dover, C. Thompson, A. Knox, J.
Plant physiology.
2001.
126(1).
210-21.
Altered cell wall disassembly during ripening of Cnr tomato fruit: implications for cell adhesion and fruit softening.
Planta (2002)
Show / hide abstract
Show / hide abstract
The Cnr ( C olourless n on- r ipening) tomato ( Lycopersicon esculentum Mill.) mutant has an aberrant fruit-ripening phenotype in which fruit do not soften and have reduced cell adhesion between pericarp cells. Cell walls from Cnr fruit were analysed in order to assess the possible contribution of pectic polysaccharides to the non-softening and altered cell adhesion phenotype. Cell wall material (CWM) and solubilised fractions of mature green and red ripe fruit were analysed by chemical, enzymatic and immunochemical techniques. No major differences in CWM sugar composition were detected although differences were found in the solubility and composition of the pectic polysaccharides extracted from the CWM at both stages of development. In comparison with the wild type, the ripening-associated solubilisation of homogalacturonan-rich pectic polysaccharides was reduced in Cnr. The proportion of carbohydrate that was chelator-soluble was 50% less in Cnr cell walls at both the mature green and red ripe stages. Chelator-soluble material from ripe-stage Cnr was more susceptible to endo-polygalacturonase degradation than the corresponding material from wild-type fruit. In addition, cell walls from Cnr fruit contained larger amounts of galactosyl- and arabinosyl-containing polysaccharides that were tightly bound in the cell wall and could only be extracted with 4 M KOH, or remained in the insoluble residue. The complexity of the cell wall alterations that occur during fruit ripening and the significance of different extractable polymer pools from cell walls are discussed in relation to the Cnr phenotype.
Orfila, Caroline. Huisman, Miranda. Willats, William. van, Alebeek. Schols, Henk. Seymour, Graham. Knox, J.
Planta.
2002.
215(3).
440-7.
Genetic analysis and FISH mapping of the Colourless non-ripening locus of tomato.
(2002)
Show / hide abstract
Show / hide abstract
Cnr ( Colourless non-ripening) is a dominant pleiotropic ripening mutation of tomato ( Lycopersicon esculentum) which has previously been mapped to the proximal region of tomato chromosome 2. We describe the fine mapping of the Cnr locus using both linkage analysis and fluorescence in situ hybridisation (FISH). Restriction fragment length polymorphism (RFLP)-, amplified restriction fragment polymorphism (AFLP)-, and cleaved amplified polymorphic sequence (CAPS)-based markers, linked to the Cnrlocus were mapped onto the long arm of chromosome 2. Detailed linkage analysis indicated that the Cnr locus was likely to lie further away from the top of the long arm than previously thought. This was confirmed by FISH, which was applied to tomato pachytene chromosomes in order to gain an insight into the organisation of hetero- and euchromatin and its relationship to the physical and genetic distances in the Cnr region. Three molecular markers linked to Cnr were unambiguously located by FISH to the long arm of chromosome 2 using individual BAC probes containing these single-copy sequences. The physical order of the markers coincided with that established by genetic analysis. The two AFLP markers most-closely linked to the Cnr locus were located in the euchromatic region 2.7-cM apart. The physical distance between these markers was measured on the pachytene spreads and estimated to be approximately 900 kb, suggesting a bp:cM relationship in this region of chromosome 2 of about 330 kb/cM. This is less than half the average value of 750 kb/cM for the tomato genome. The relationship between genetic and physical distances on chromosome 2 is discussed.
Tör, M.. Manning, K.. King, G.. Thompson, A.. Jones, G.. Seymour, G.. Armstrong, S..
.
2002.
104(2-3).
165-170.
Effect of the Colorless non-ripening mutation on cell wall biochemistry and gene expression during tomato fruit development and ripening.
Plant physiology (2004)
Show / hide abstract
Show / hide abstract
The Colorless non-ripening (Cnr) mutation in tomato (Solanum lycopersicum) results in mature fruits with colorless pericarp tissue showing an excessive loss of cell adhesion (A.J. Thompson, M. Tor, C.S. Barry, J. Vrebalov, C. Orfila, M.C. Jarvis, J.J. Giovannoni, D. Grierson, G.B. Seymour [1999] Plant Physiol 120: 383-390). This pleiotropic mutation is an important tool for investigating the biochemical and molecular basis of cell separation during ripening. This study reports on the changes in enzyme activity associated with cell wall disassembly in Cnr and the effect of the mutation on the program of ripening-related gene expression. Real-time PCR and biochemical analysis demonstrated that the expression and activity of a range of cell wall-degrading enzymes was altered in Cnr during both development and ripening. These enzymes included polygalacturonase, pectinesterase (PE), galactanase, and xyloglucan endotransglycosylase. In the case of PE, the protein product of the ripening-related isoform PE2 was not detected in the mutant. In contrast with wild type, Cnr fruits were rich in basic chitinase and peroxidase activity. A microarray and differential screen were used to profile the pattern of gene expression in wild-type and Cnr fruits. They revealed a picture of the gene expression in the mutant that was largely consistent with the real-time PCR and biochemical experiments. Additionally, these experiments demonstrated that the Cnr mutation had a profound effect on many aspects of ripening-related gene expression. This included a severe reduction in the expression of ripening-related genes in mature fruits and indications of premature expression of some of these genes in immature fruits. The program of gene expression in Cnr resembles to some degree that found in dehiscence or abscission zones. We speculate that there is a link between events controlling cell separation in tomato, a fleshy fruit, and those involved in the formation of dehiscence zones in dry fruits.
Eriksson, EM. Bovy, A. Manning, K. Harrison, L. Andrews, J. De Silva, J. Tucker, GA. Seymour, GB.
Plant physiology.
2004.
136(4).
4184-97.
A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening.
Nature genetics (2006)
Show / hide abstract
Show / hide abstract
A major component in the regulatory network controlling fruit ripening is likely to be the gene at the tomato Colorless non-ripening (Cnr) locus. The Cnr mutation results in colorless fruits with a substantial loss of cell-to-cell adhesion. The nature of the mutation and the identity of the Cnr gene were previously unknown. Using positional cloning and virus-induced gene silencing, here we demonstrate that an SBP-box (SQUAMOSA promoter binding protein-like) gene resides at the Cnr locus. Furthermore, the Cnr phenotype results from a spontaneous epigenetic change in the SBP-box promoter. The discovery that Cnr is an epimutation was unexpected, as very few spontaneous epimutations have been described in plants. This study demonstrates that an SBP-box gene is critical for normal ripening and highlights the likely importance of epialleles in plant development and the generation of natural variation.
Manning, Kenneth. Tör, Mahmut. Poole, Mervin. Hong, Yiguo. Thompson, Andrew. King, Graham. Giovannoni, James. Seymour, Graham.
Nature genetics.
2006.
38(8).
948-52.
Genomic organization, phylogenetic comparison and differential expression of the SBP-box family of transcription factors in tomato.
Planta (2011)
Show / hide abstract
Show / hide abstract
SBP-box genes represent transcription factors ubiquitously found in the plant kingdom and recognized as important regulators of many different aspects of plant development. In this study, 15 SBP-box gene family members were identified in tomato and analyzed with respect to their genomic organization and other structural features. Phylogenetic reconstruction based on the DNA-binding SBP-domain, allowed the classification of the SlySBP proteins into eight groups representing clear orthologous relationships to family members of other flowering plants and the moss Physcomitrella. In order to have a better understanding of their possible function in the development of a fleshy-fruit species like tomato, the mRNA expression levels of all SlySBP genes were quantified in vegetative and reproductive organs of plants, at different stages of growth. As transcripts of ten SlySBP genes were found to carry putative miR156- and miR157-response elements, the expression levels of the corresponding microRNAs were determined as well, revealing different patterns of expression. In addition, eight putative miR156 and four miR157 encoding loci could be identified in the tomato genome, four of them forming a polycistronic cluster. Whereas miR156 and miR157 levels were highest in seedlings, leaves and anthers of young flowers, most miR156-targeted SlySBP genes were found to be expressed in young inflorescences and during fruit development and ripening, suggesting a particularly important role during tomato reproductive growth. The data presented provide a basis for future clarification of the various functions that SBP-box gene family members play in tomato growth and development.
Salinas, M. Xing, S. Höhmann, S. Berndtgen, R. Huijser, P.
Planta.
2011.
().
.
 Ontology annotations (4) Ontology annotations (4)
 Ontology annotations (4) Ontology annotations (4)
| [Add ontology annotations] |
[loading...]
 Related views Related views
 Related views Related views
|
- Genomic details
| User comments |
Please wait, checking for comments. (If comments do not show up, access them here)
Your Lists
Public Lists
List Contents
List Validation Report: Failed
Elements not found:
Optional: Add Missing Accessions to A List
Mismatched case
Click the Adjust Case button to align the case in the list with what is in the database.
Multiple mismatched case
Items listed here have mulitple case mismatches and must be fixed manually. If accessions need to be merged, contact the database directly.
List elements matching a synonym
Multiple synonym matches
Fuzzy Search Results
Synonym Search Results
Available Seedlots
Your Datasets
Public Datasets
Dataset Contents
Dataset Validation Failed
Elements not found:
Your Calendar
Having trouble viewing events on the calendar?
Are you associated with the breeding program you are interested in viewing?
Add New Event
Event Info
| Attribute | Value |
|---|---|
| Project Name: | |
| Start Date: | |
| End Date: | |
| Event Type: | |
| Event Description: | |
| Event Web URL: |
Edit Event
Login
Forgot Username
If you've forgotten your username, enter your email address below. An email will be sent with any account username(s) associated with your email address.
Reset Password
To reset your password, please enter your email address. A link will be sent to that address with a link that will enable you to reset your password.
Create New User
Working